Example 1. SARS Main Protease
I. Searching the structure templateII. Sequence Alignment using BioEdit
III. Adjusting the Alignment manually in SWISS-PDBViewer
IV. Model Building
Computer programs used in this example
1. SWISS-PDBViewer
2. BioEdit
I. Searching the structure template
1. Prepare the primary sequence of SARS Main Protease in FASTA format:3cpro.fas
>SARS_3CPro
SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDTVYCPRHVICTAEDML
NPNYEDLLIRKSNHSFLVQAGNVQLRVIGHSMQNCLLRLKVDTSNPKTPK
YKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNHTIKGSFLNGSCGSVGF
NIDYDCVSFCYMHHMELPTGVHAGTDLEGKFYGPFVDRQTAQAAGTDTTI
TLNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYEPLTQDHVDIL
GPLSAQTGIAVLDMCAALKELLQNGMNGRTILGSTILEDEFTPFDVVRQC
SGVTFQ
2. Double click "spdbv.exe" to run the program SWISS-PDBViewer.
3. Load primary sequence "3cpro.fas" by clicking "SwissModel -> Load Raw Sequence to Model ..."
You can inspect the sequence by "Window -> Alignment". After loading the sequence, you will see an extended polypeptide chain.
4. You can search for the appropiate template structure by "SwissModel -> Find appropiate ExPDB template".
Press "Submit".
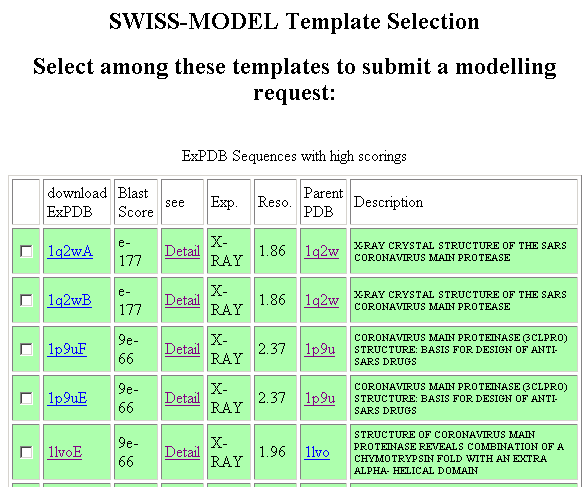
The top 2 hits (1q2wA and 1q2wB) are in fact the structure of SARS Main Protease solved by X-ray crstallography. The 1p9u and 1lvo is the structure of main protease (with and without inhibitor) of another coronavirus (Transmissible gastroenteritis virus). In this example, we select 1p9uF as the template.
5. Load 1p9uF.pdb to SWISS-PDBViewer by "File -> Open PDB File ...". You will be able to see two structures in the main window and two sequences in the alignment window. Before you can predict the structure, you will need to obtain a good alignment between the sequences of main protease of SARS virus and TGV.
II. Sequence Alignment using BioEdit
1. Sequence Alignment is the most important step in homology modelling. Incorrect alignment will lead to wrong 3D model. Alignment between two sequences can be obtained by pairwise alignment method (e.g. BLAST). However, in most cases alignment can be improved using the technique of multiple sequence alignment.2. The main protease is found in the genome of all coronaviruses. The file 3cpro_all.fas contain the sequence of main protease of related coronaviruses.
Double click the file 3cpro_all.fas to open it:
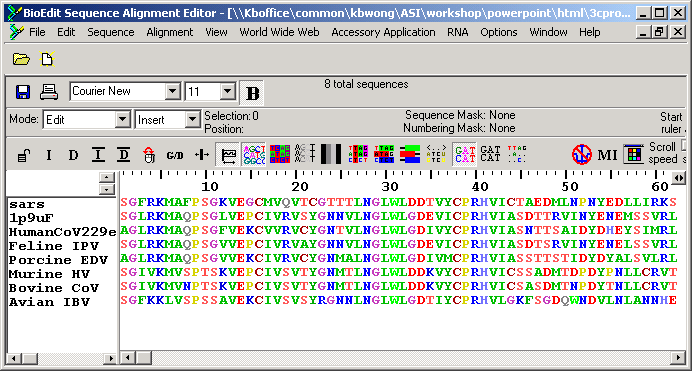
3. You can perform multiple sequence alignment by "Accessory Application -> ClustalW Mutliple Alignment". Use default setting and press the button "Run ClustalW"
Notice the sequences have been aligned accordingly:
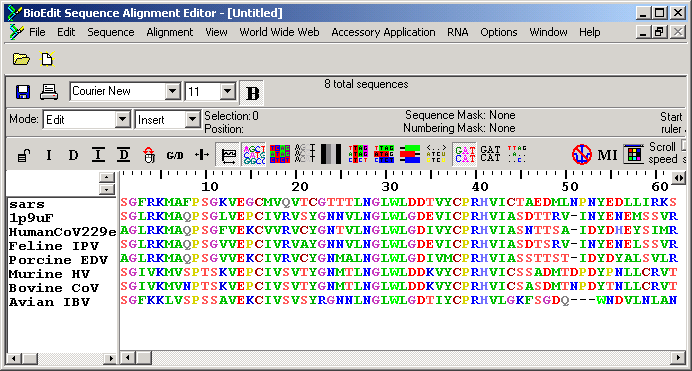
The sequence identity between SARS and TGV Main Protease is 43%, which indicates that the structure prediction is likely to be successful.
4. The alignment can be exported to RTF format by "File -> Graphic View", then Click "Export as Rich Text". After we have obtained the sequence alignment, we can now go back to SWISS-PDBViewer.
III. Adjusting the
Alignment manually in SWISS-PDBViewer
1. Inspect the Alignment windows and compare it with the alignment
obtained in "BioEdit". For example, P-50 of SARS protease should align
with I-51 of TGV protease.
To adjust the alignment manually, first click on residue I-51 of 1p9uF and then type spacebar:

| insert a gap | space bar |
| delete a gap | backspace |
| left/right arrows | move the selected residues to the left/right |
2. Can you adjust the alignment until it matches the alignment obtained from "BioEdit"?
3. SWISS-PDBViewer can read and write alignment in the FoldFit format. The alignment file 3cpro.foldfit.txt is included in case you get lost with the alignment.You can load the correct alignment to SWISS-PDBViewer by "SwissModel -> Load FoldFit Alignment ...".
IV. Model Building
1. In this tutorial, the predicted model is built by the SWISS-MODEL server. You can submit the job to the server by clicking "SwissModel -> Submit modelling request ...". When prompted, just type a file name (e.g. 3cpro.htm) and you will see:
2. Typing your email address and the file name of the SWISS-MODEL project file, then press the button "Send Request". The results will be sent to you by email.
3. If you can't check your email, the resulting pdb file can be found here.
V. Exercise
Predict the structure of main protease of Avian infectious bronchitis virus. The primary sequence of the protein is:>AIBV_3CPro
SGFKKLVSPSSAVEKCIVSVSYRGNNLNGLWLGDTIYCPRHVLGKFSGDQWNDVLNLANNHEFEVTTQHG
VTLNVVSRRLKGAVLILQTAVANAETPKYKFIKANCGDSFTIACAYGGTVVGLYPVTMRSNGTIRASFLA
GACGSVGFNIEKGVVNFFYMHHLELPNALHTGTDLMGEFYGGYVDEEVAQRVPPDNLVTNNIVAWLYAAI
ISVKESSFSLPKWLESTTVSVDDYNKWAGDNGFTPFSTSTAITKLSAITGVDVCKLLRTIMVKNSQWGGD
PILGQYNFEDELTPESVFNQIGGVRLQ