Computer Modeling of Biomolecules
Presented by
Kam Bo Wong
Department of Biochemistry & Molecular Biotechnology Programme
email: kbwong@cuhk.edu.hk
http://smart.bch.cuhk.edu.hk/kbwong/index.htm
4. Base-pairing in DNA
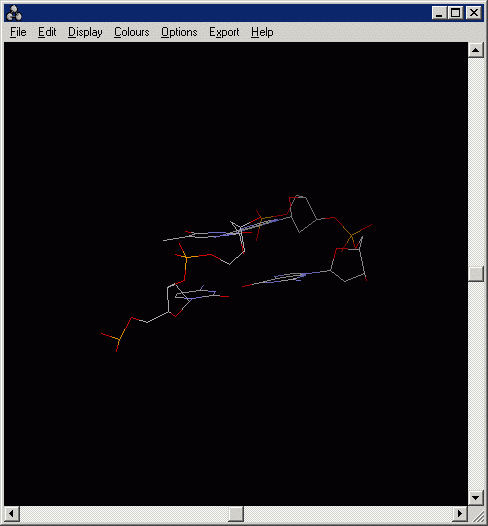
4.1 Load the "basepair.pdb".
The structure represents a short fragment of DNA consisting
of 2 base-pairs. One of them is an A-T pair and the other one is a G-C pair.
Can you identify which one is which?
4.2 Activate the command line interface of RasMol by clicking the "RasMol Command Line Button" on the Toolbar.
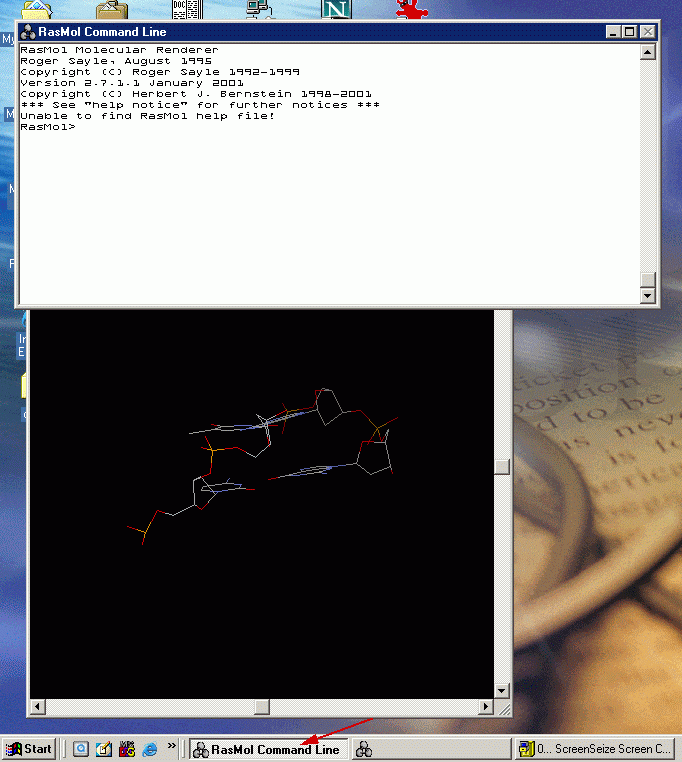
4.3 Display the hydrogen bond pattern
The command line interface allows you to access the more
advanced features of RasMol. One of them is to show hydrogen bonding pattern.
Try typing the command "hbond on" and you should see the hydrogen
bonding pattern between GC and AT base pairing.
In the subsequent tutorial, we will use the following to represent typing command in the command line interface:
RasMol> hbond on
Given that a G-C pairing involves formation of three hydrogen bonds and a
A-T pairing involves two hydrogen bonds, can you now identify which one is a
G-C pair and which one is A-T pair?
· Can you figure out how to turn off
the hydrogen bond display?
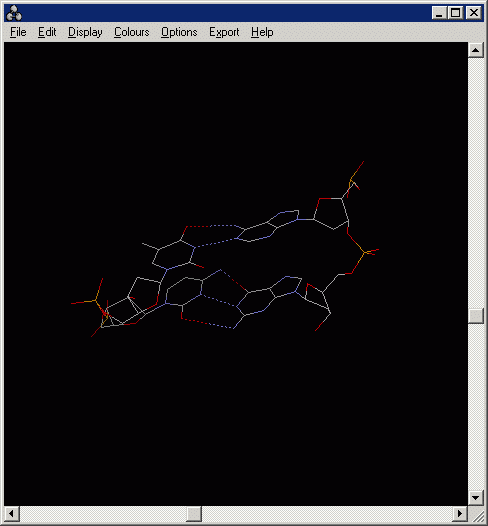
4.4 Measuring the distance between two atoms.
To turn on distance monitor:
RasMol> set picking monitor
Then pick two atoms in the Main Windows of RasMol:
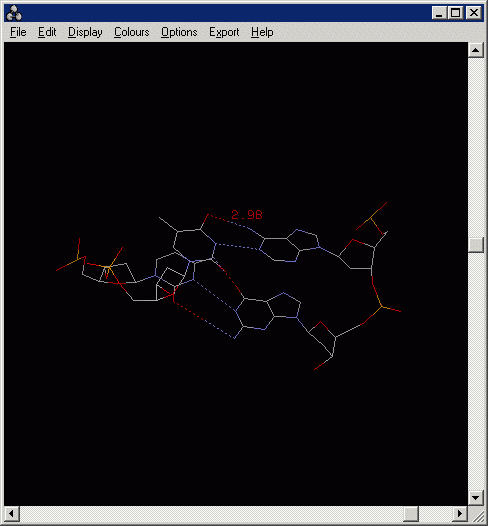
In the above example, the distance between A-T pair is 2.98 Angstrom (i.e.
0.298 nm)
To turn off distance monitor and set the picking mode back to default:
RasMol>
monitor off
RasMol> set
picking ident