
Teacher Development Course
Using molecular modeling software to view protein structures
Presented by Kam Bo Wong
Department of Biochemistry & Molecular Biotechnology Programme
email: kbwong@cuhk.edu.hk
http://smart.bch.cuhk.edu.hk/kbwong/index.htm
2. Using Chime to look at a small organic molecule.
After you have installed Chime, your browser are now ready to view
3D structure of molecules. We will first look at a small molecule: urea.
The 3D structural information of a molecule is stored in a so-called 'PDB'
format. The 3D structure file for urea is named 'urea.pdb' (remember the
file must have a '.pdb' extension)
2.1 You can now view the structure of urea by open the file or
just simply click here.

2.2 You can access the menu interface of Chime by clicking the
right mouse button:
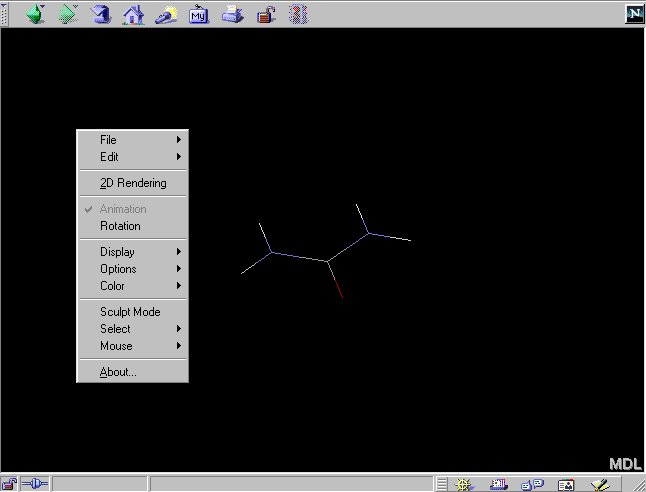
Changing the display mode in Chime
2.3 The default display mode is 'wireframe' where only bond between
atoms are shown. You can change the display mode to 'Ball-and-stick' by:
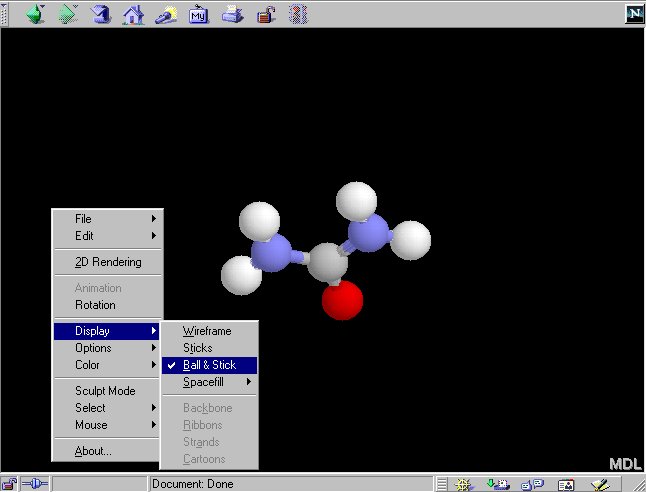
In 'Ball-and-stick' model, atoms are represented by spheres and bonds
are represented by thicker lines.
2.4 You can also change the display mode to 'Spacefill':
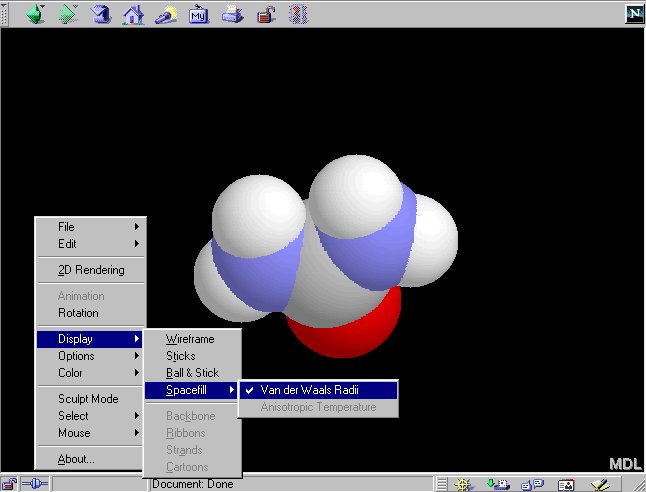
In 'Spacefill' model, atoms are represented by spheres with sizes dependent
on their atom types (e.g. hydrogen atoms are smaller than the nitrogen
atoms.) In the default color setting, hydrogen atoms are white,
nitrogen atoms are blue and oxygen atoms are
red.
2.5 Mouse Control in Chime.
-
Rotate the molecule - Press the left mouse
button and dragging the mouse.
-
Move the molecule - Press the 'CTRL' key AND
the right mouse button.
-
Zoom in/out - Press the 'SHIFT key AND the
left
mouse button.





