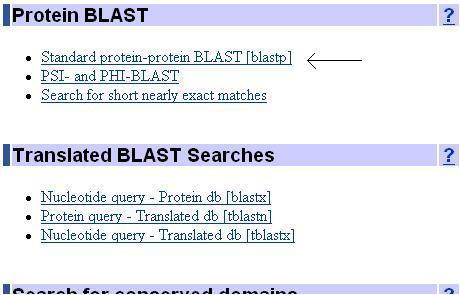
Protein BLAST allows one to input protein sequences and compare these against other protein sequences. We will introduce one of the protein blast called Standard protein-protein BLAST.
Standard protein-protein BLAST allow you to takes protein sequences in FASTA format, GenBank Accession numbers or GI numbers and compares them against the NCBI protein databases.
First please go to the homepage of BLAST. To decode the amino acid sequence to a known protein, Standard protein-protein BLAST is used. Search for protein blast and click the Standard protein-protein BLAST.

You can paste the protein sequence in the space of search, and you can choose database, such as rat, drosophila (normally we use nr for human genome). Then click blast!.
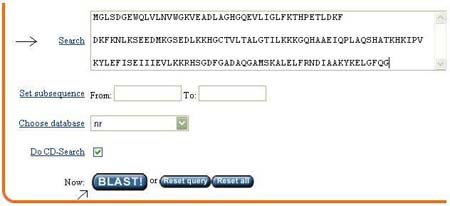
Then you will see a picture in the page, it shows the domain present in the protein. You can click the picture for more details (You can view the 3D structure of the protein there). To continue blast, click Format!.

Then a new browser which will automatically update will be opened. Wait for a while until the search finishes, and the result will be displayed in the new browser.

¡@
When the result is displayed, see the picture of distribution carefully. Click on the bar which makes the greatest alignment, this protein sequence is most similar to the query protein sequence.

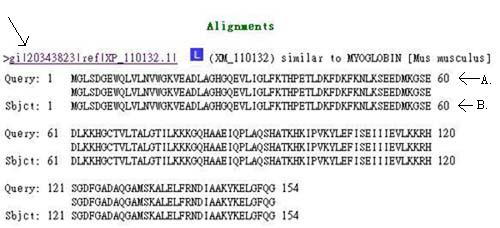
In the alignment section, you can see how much your query sequence (A.) matches the sequence in the database (B.). You can click on the hyperlink for more details of the protein.
¡@