|
Research Interests
- Structure determination of proteins by NMR spectroscopy and
X-ray
crystallography
- Protein engineering and design
- Simulation and modeling of protein
Research
Projects
- Structure
basis of thermostability of proteins
- Structure-function
studies of acidic ribosomal proteins
- How H. pylori urease matures?
- Substrate specificity and inhibitor
design for 3C-like protease of Coronavirus
Structure
basis of thermostability of proteins
Understanding the 'rules' of structural adaptation of how proteins
remain stable and active at high temperatures is not only of great
academic interests but also has potential applications in
biotechnology. For example, engineering of thermostable industrial
enzymes offers the benefits of increased rate of chemical reactions at
higher temperatures. At high temperatures, inactivation of proteins is
often caused by unfolding, which exposes the polypeptide chains to
various irreversible processes (e.g. chemical modification and
aggregation). Proteins from thermophilic organisms, which
adapt to grow at elevated temperatures (55-110�C), are
excellent models for this kind of studies. Although some intracellular
factors are reported to stabilize protein
in vivo, most thermophilic proteins are intrinsically more stable at
high temperatures. Learning how these thermophilic proteins achieve
thermostability will provide rules for rational design of thermostable
proteins.
In our laboratory, we are currently
working on two model proteins to study the structural basis of protein
thermostability:
(1) Ribosomal protein
L30e from Thermococcus
celer
(2) Acylphosphatase from Pyrococcus
horikoshii

Collaborators
Mark
Bycroft, Centre for Protein Engineering, MRC, Cambridge, UK
George
Makhatdze, Penn. State University, USA
Kong-Hung Sze, University of Hong Kong
Selected Publications
- Chan, S.H., Wilbanks, C.C., Makhatadze, G.I., Wong, K.B. (2012) Electrostatic
Contribution of Surface Charge Residues to the Stability of a
Thermophilic Protein: Benchmarking Experimental and Predicted pKa Values, PLoS One 7, e30296.
- Lam, S. Y., Yeung, R. C. Y., Yu, T.-H., Sze, K.-H., and Wong, K.-B. (2011) A
Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at
High Temperatures at the Expense of Low-Temperature Activity, PLoS Biology 9, e1001027.
- Chan C.H., Yu, T.H., Wong, K.B. (2011) Stabilizing salt-bridge enhances protein thermostability by reducing the heat capacity change of unfolding. PLoS One 6, e21624.
- Lee
CF, Makhatadze GI, Wong KB: Effects
of charge-to-alanine substitutions on the stability of ribosomal
protein L30e
from Thermococcus celer. Biochemistry
2005, 44:16817-16825.
- Lee CF, Allen MD, Bycroft M,
Wong KB: Electrostatic
interactions contribute to reduced heat capacity change of unfolding in
a thermophilic
ribosomal protein L30e. J Mol Biol 2005,
348:419-431.
- Cheung YY, Lam SY, Chu WK,
Allen MD, Bycroft M, Wong KB: Crystal
structure of a hyperthermophilic
archaeal acylphosphatase from Pyrococcus horikoshii--structural
insights into
enzymatic catalysis, thermostability, and dimerization. Biochemistry 2005, 44:4601-4611.
- Wong KB, Lee CF, Chan SH, Leung
TY, Chen YW, Bycroft M: Solution structure
and thermal stability of
ribosomal protein L30e from hyperthermophilic archaeon Thermococcus
celer. Protein Sci 2003,
12:1483-1495.
- Chen YW, Bycroft M, Wong KB: Crystal
structure of ribosomal protein L30e from the extreme thermophile
Thermococcus
celer: thermal stability and RNA binding. Biochemistry
2003, 42:2857-2865.
Press release
Structure-functions
of ribosomal
stalk proteins and its interactions with ribosomal toxin
Acidic ribosomal proteins, P0, P1, and P2, are the
constituent of the P-complex that forms the stalk of eukaryotic
ribosome, which involves in binding of translation factors and their
activation by GTP-hydrolysis. The ribosomal stalk proteins are also
involved in interacting with ribosome-inactivating proteins.
We are interested to determine the
structure of acidic protein proteins, which will complement recent
structural studies of prokaryotic and archaeal ribosomes, and will
contribute to a better understanding of structure-function of
eukaryotic ribosome.
(1) Solution structure of the N-terminal dimerization domain of P2 (Lee
et al., 2010)
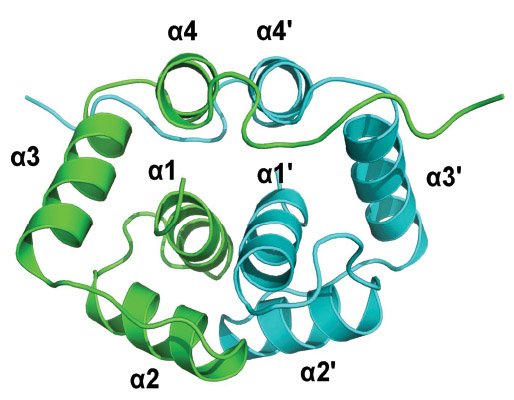
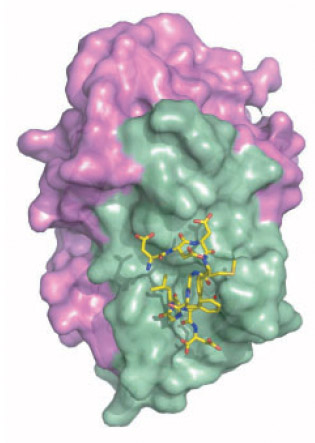
(2) Crystal structure of C-terminal conserved region of P2 in complex
with trichosanthin (Too et al., 2009)
Collaborators
Pang-Chui
Shaw, Chinese University of Hong Kong
Guang Zhu, Hong Kong
University of Science and Technology
Kong-Hung Sze, University of Hong Kong
Selected Publication
- Lee KM, Yusa K, Chu LO, Yu, CWH, Oono M, Miyoshi T, Ito K, Shaw PC, Wong KB, Uchiumi T. (2013) Solution
structure of human P1-P2 heterodimer provides insights into the role of
eukaryotic stalk in recruiting ribosome-inactivating protein
trichosanthin to the ribosome. Nucleic Acids Res, 41, 8776-8787
- Lee, K.M., Yu, W.H., Chiu, Y.H., Sze, K.H., Shaw, P.C.,
Wong, K.B. (2012) Solution structure of the dimerization domain of the
eukaryotic stalk P1/P2 complex reveals the structural organization of
eukaryotic stalk complex, Nucleic Acids Res 40:3172-82
- Lee KM, Yu CW, Chan DS, Chiu
TY, Zhu G, Sze KH, Shaw PC, Wong KB: Solution
structure of the dimerization
domain of ribosomal protein P2 provides insights for the structural
organization of eukaryotic stalk. Nucleic
Acids Res 2010, 38:5206-5216.
- Law SK, Wang RR, Mak AN, Wong
KB, Zheng YT, Shaw PC: A switch-on mechanism
to activate maize ribosome-inactivating protein
for targeting HIV-infected cells. Nucleic
Acids Res 2010, 38:6803-6812.
- Too PH, Ma MK, Mak AN, Wong YT,
Tung CK, Zhu G, Au SW, Wong KB, Shaw PC: The
C-terminal fragment of the ribosomal P
protein complexed to trichosanthin reveals the interaction between the
ribosome-inactivating protein and the ribosome. Nucleic Acids Res 2009, 37:602-610.
- Mak AN, Wong YT, An YJ, Cha SS,
Sze KH, Au SW, Wong KB, Shaw PC: Structure-function
study of maize
ribosome-inactivating protein: implications for the internal
inactivation
region and the sole glutamate in the active site. Nucleic Acids Res 2007, 35:6259-6267.
- Chan DS, Chu LO, Lee KM, Too
PH, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB: Interaction
between trichosanthin, a ribosome-inactivating
protein, and the ribosomal stalk protein P2 by chemical shift
perturbation and
mutagenesis analyses. Nucleic Acids
Res 2007, 35:1660-1672.
How H. pylori urease matures?
Helicobacter pylori is
the only bacterium known to thrive in the human stomach. It damages the
mucous coating of the gut, allowing stomach acid to eat away the
sensitive organ lining and causing ulcers. H. pylori produces
urease to spur the breakdown of urea, a naturally occurring chemical in
the body, so that urea can release ammonia to neutralize the acidic
environment in the gut, allowing pathogens to thrive. Unlike most other
enzymes, urease does not work immediately after being produced by the
bacterium. Two nickel ions have to be delivered to activate it. Our
work focuses on four urease maturation factors, UreE, UreF, UreH, UreG
that help the activation of urease: UreE, UreF, UreG and UreH.
Crystal structure of urease maturation factors UreF/UreH complex (Fong et al., 2011)
Collaborators
Yu Wai Chen, King's College London
HZ Sun, University of Hong Kong
Selected Publication
- Fong, Y.H., Wong, H.C., Chuck, C.P., Chen, Y.W., Sun, H., Wong, K.B. (2011)
Assembly of the preactivation complex for urease maturation in
Helicobacter pylori: Crystal Structure of the UreF/UreH complex, J. Biol. Chem. 286, 43241. (selected as paper of the week)
-
Fong YH, Wong HC, Yeun MH, Lau PH, Chen YW, Wong KB. (2013) Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLoS Biol, 11, e1001678.

Press release
Substrate
specificity and inhibitor design for 3C-like protease of Coronavirus
Coronavirus (CoV) infections cause many respiratory tract diseases like
common cold, bronchiolitis and pneumonia in human. The most serious
outbreak was caused by the severe acute respiraory syndrome (SARS)
virus in 2003 that has resulted in a death toll of more than 700
world-wide. Although the SARS outbreak was brought under control, the
high mutation and recombination rate, and a history of interspecies
transmission suggest that coronavirus infections remain a potential
threat to public health. As there is no approved drug currently
available for CoV infections, we see there is a need to develop novel
drug to combat against future outbreak of CoV infection. The 3C-like
protease, which is present in all CoV and play vital role in viral
replication, is an ideal drug target for this purpose. Our group is
interested in profiling of the substrate specificity and in a rational
design of peptidomimetic inhibitors for the 3C-like protease.
Histidine is allowed at P1 position of 3C-like protease of SARS-CoV (Chuck CP
et al., 2010)
Collaborators
Hak-fun Chow, Chinese University of Hong Kong
David C.C. Wan, Chinese University of Hong Kong
Selected Publication
- Chuck CP, Chen C, Ke Z, Wan DC, Chow HF, Wong KB. (2013) Design,
synthesis and crystallographic analysis of nitrile-based broad-spectrum
peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur J Med Chem, 59:1-6.
- Chuck, C.P., Chow, H.F., Wan, C.C., Wong, K.B. (2011) Profiling of Substrate Specificities of 3C-Like Proteases from Group 1, 2a, 2b, and 3 Coronaviruses, PLoS One, 6, e27228
- Chuck CP, Chong LT, Chen C, Chow HF, Wan
DC, Wong KB: Profiling of substrate
specificity of SARS-CoV 3CL. PLoS
One 2010, 5:e13197.
| |

