4. Database of 3D protein structures
The three dimensional structure of yeast ribosomal protein L30 is known. We will take a look at the 3D structure of the protein and check if our previous secondary structure prediction agrees with the experiments. All 3D structures of proteins are stored in the Protein Data Bank (PDB).
1. Go to the Protein Data Bank. Search the database with 'yeast ribosomal protein l30'. To simplify result, check the “remove sequence homologs”.
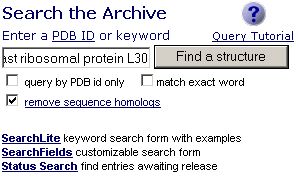
Search Results:
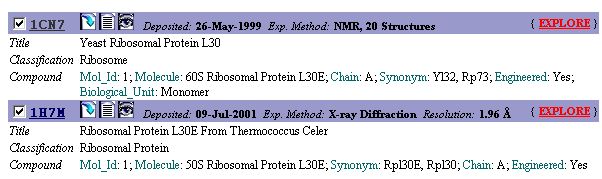
All entries in the PDB is identified by a four-letter code. The PDB code for
the yeast ribosomal protein L30 is '1CN7'. Note that 1H7M code for
another L30e protein from Thermococcus celer.
Select '1CN7' and press the 'Explore' button.
2. You will see the following window. Click 'Sequence Details' to view the secondary structure assignment of this protein.
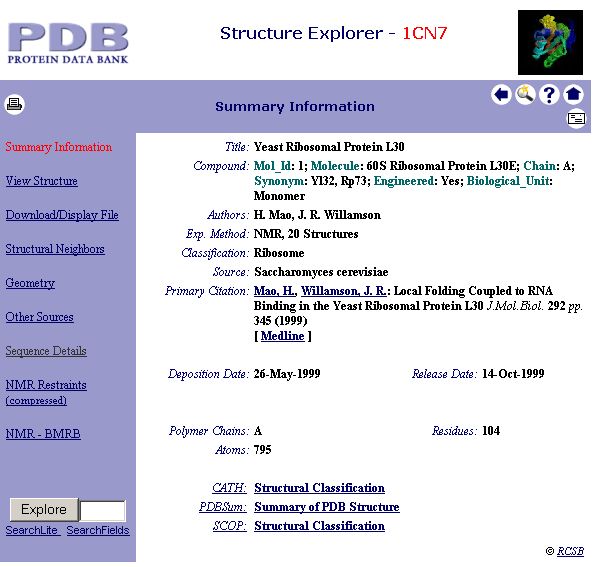
3. Scroll down the window until you see:

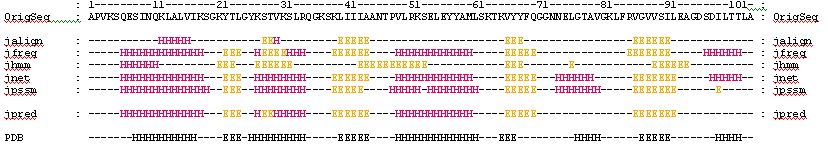
4. Your can check the secondary structure of the protein to see if our previous prediction agrees with the experiment:
Consider only the Alpha-Helices (H) and Beta-Strands (E):
5. Now we want to look at the structure of this protein. Go to 'View Structure'.
6. Click 'Ribbons (500x500)' to view a still image of the structure. Or click here if the internet is too slow.
7. You can also look at the 3D view by clicking the 'QuickPDB' button.
Can you identify the secondary
structure elements of this protein?