3. Secondary structure prediction
APVKSQESIN
QKLALVIKSG KYTLGYKSTV KSLRQGKSKL IIIAANTPVL
RKSELEYYAM LSKTKVYYFQ
GGNNELGTAV GKLFRVGVVS ILEAGDSDIL
TTLA
1. Go to the Jpred
server
2. Enter your email address in Box 1.
3. Copy the query sequence and paste it into the window:
APVKSQESIN
QKLALVIKSG KYTLGYKSTV KSLRQGKSKL IIIAANTPVL
RKSELEYYAM
LSKTKVYYFQ GGNNELGTAV GKLFRVGVVS ILEAGDSDIL
TTLA
4. Click the option to bypass an automatic scan of the current Brookhaven Protein Database.
You need to do this because the three-dimensional structure of the ribosomal
protein L30 is known. By default, Jpred will not perform any secondary
structure prediction unless you force it to do so.

5. Press the "Run secondary structure prediction" button.
The prediction takes very long to run. To save time, the instructor has run the prediction for you and you can view the result here.
6. Scroll the result window until you see:
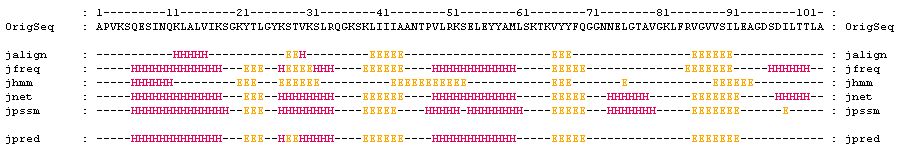
Alpha-helices are represented by the letter 'H' while beta-strands are
represented by the letter 'E'. The prediction suggested the ribosomal protein
consists of 5 alpha-helices and 4 beta-strands.